Reconstructive Connectomics
Cryonics, July 2013
by Chana Phaedra
“If we are acquainted with the principles upon which this mechanism [the brain] operates, we may infer its function from its structure, regarding the former as a natural outcome of the latter.” — Theodor Meynert, 1885
If you’ve been paying attention to the latest initiatives in neuroscience you likely have heard something about the “connectome.” You may be wondering what the connectome is and what all the hubbub is about. This article seeks to briefly define the connectome, describe the goals of connectomics, and make a case for using what we learn (and what we already know) from connectomics to pursue meaningful cryonics resuscitation research today.
The connectome is a comprehensive description of how neurons and brain regions are interconnected. It is commonly conceptualized as a sort of “wiring diagram” of the brain. Connectomics, then, is the branch of biotechnology concerned with applying the techniques of image acquisition and analysis to the structural mapping of sets of neural circuits, with organizing the results in databases, and with applications of the data.
Motivating modern research in this area is the hypothesis that mapping of anatomical circuits represents a fundamental step toward understanding brain function and physiology. Until now, functional studies of cells and circuits have largely been carried out in the absence of detailed and specific information about the underlying connectivity.
Connectome mapping can be carried out directly, by deploying anatomical techniques, or indirectly, by attempting to infer connections between circuit elements based on their temporal dynamics or functional responses. Besides increased general knowledge, the most obvious application of connectomics data includes the treatment of disease, as in neurology or fundamental neuroscience (i.e., to learn more about the role of connectional disturbances in brain dysfunction and disease).
Another application or goal of connectomics can be to repair damage associated with cryopreservation with the intention of patient resuscitation. This kind of “reconstructive connectomics” (a phrase coined by Aschwin de Wolf in Cryonics May 2013) would be a subdiscipline of the field of connectomics that studies the pathological changes of neural connections in the brain with the aim of in silico (i.e., computer-aided) repair.
Reconstructive connectomics is the modern-day realization of what Thomas Donaldson termed “neural archeology,” a concept described in detail in his 1987 article of the same name. In general terms, Donaldson equates the task ahead of cryonicists with that encountered by traditional archeologists. Though space limits our ability to consider this prescient article in full, let us look at a most illuminating section:
“The first thing done in examining an archeological site is to carefully plot the relation of all the fragments to one another. Debris has a structure too. We discover this structure by looking at the relations of its parts to one another, not just by looking at the parts. (Archaeologists in Central America complain constantly that valuable artifacts are taken away and sold, with no record of where they were found, in relation to what.) If a protein has two degradation parts, we can learn a lot by knowing where these parts are found in the remains of a cell.
“In fact, one way of looking at cryonics is that it is simply a way of making such a detailed record. Here is a patient’s brain, in the condition it was when we lost him.”
Indeed, many fields use reconstructive techniques—taking a limited amount of information from a system and extrapolating what is known about that system to “fill in the gaps” and reconstruct the original system state. For example, facial reconstruction is performed in forensics and archeology. Mathematical and scientific modeling is also used in genomics and bioinformatics.
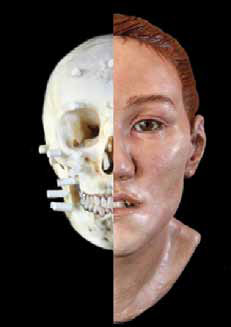
Facial reconstruction provides a good example of inferring an original structure from incomplete information.
Inherent to Donaldson’s statement above is the fact that some amount of damage has been inflicted upon the brain of the cryonics patient. Indeed, while repair is not intrinsic to cryonics, most cryonics patients will require repair. However, one of the biggest myths in cryonics today is that not much of interest can currently be said about repair, let alone that repair efforts can begin now.
I will argue otherwise. In the context of resuscitating a cryopreserved patient, we must answer three important questions: (1) What needs to be repaired? (2) What repair strategies can be utilized? and (3) What level of information resolution is sufficient? By characterizing the amount and type of damage in detail and linking those forms of damage to contemporary or envisioned repair strategies, especially making use of information obtained from connectomics research, we can begin the process of reconstructing the functional person from the preserved brain, whatever state that brain may be in at the time of preservation.
Some people may believe that scanning of the patient’s damaged brain and in silico repair is sufficient for meaningful resuscitation (i.e., mind uploading). I recognize this point but remain agnostic. Let us work from the assumption that repair will be on the biological brain with the help of computer modeling.
Even if we do not have the biological or mechanical cell repair technologies that would be required for repair of the brain at the molecular level, using techniques in connectomics we can do things like simulate a specific kind of damage (e.g., ice formation, ischemia) and create 3D neural wiring maps that can be compared to controls.
Often this is not even necessary because we understand the universal language of biology and if we observe damage (e.g., a ruptured cell membrane), we know how it is supposed to look. In fact, using our knowledge of what a brain should look like, and how they should function, can get us surprisingly far. Observe, for instance, the following electron micrographs (EMs) produced by my research company, Advanced Neural Biosciences. Figure 1 is an EM of normal, undamaged rat cortical tissue (i.e., the “control” sample), while Figure 2 is an EM at the same magnification of rat cortical tissue that has suffered 21 hours of warm ischemia.

Figure 1. A control cortical electron micrograph of the rat brain.

Figure 2. Cortical electron micrograph of the rat brain after 21 hours of warm ischemia. Structure and organelles can still be clearly identified.
It is remarkable that in both the control and the 21 hour sample myelin sheaths can be easily identified. If you look at these images closely you will observe deformed/ruptured organelles and evidence of disintegration in the 21 hour group, but for many structures: (a) we know how they should look, and (b) memory and identity are not encoded in individual organelles such as mitochondria. There is substantial information still present in this damaged system to utilize in reconstructing the original state.
Much work has already been done in characterizing damage in cryonics. In brief, damage falls into the following categories: damage incurred prior to cryopreservation (i.e., “pre-mortem damage”), cerebral ischemia, cryoprotectant toxicity, ice formation, chilling injury, and dehydration. The question of utmost importance in considering these forms of damage is whether we should expect any of them to destroy (our ability to reconstruct) the connectome.
I think not.
Computational limitations currently constrain the scale and complexity at which we can do these reconstructions, but it is not necessary to do reconstructive connectomics in a human-sized brain to obtain a much greater understanding of the mechanisms of damage, the type of repair required, and the empirical content of concepts like information-theoretic death.
An open question is whether the connectome will provide sufficient information to enable resuscitation from cryopreservation. One could argue that we may need to preserve detailed information at even higher resolution (e.g., the “synaptome,” ion channels, microtubules, neurotransmitters, etc.). That the connectome is sufficient to reconstruct individual identity and memory is an assumption of mine; however, if other biochemical mechanisms are involved, similar reasoning can be applied.
It is important to keep in mind that connectome data exists at multiple scales (micro vs. macro). Very little progress has been made to date in merging or crossreferencing of connectome data across scales—for example, relating connection data from networks at cellular resolution to large-scale projections and pathways. The development of multi-modal registration tools and connectivity-based data formats for diffusion and functional MRI provides a potential model for future integration efforts across scales.
So what kind of resuscitation research can be undertaken NOW? To start, we can better characterize damage and changes to the brain incurred prior to and as a result of cryopreservation. We can then use computer modeling to reconstruct the original state in silico. Additionally, we can utilize the tools of synthetic biology to develop biological cell repair technologies.
A specific project that is currently being undertaken by Advanced Neural Biosciences is to compare control (i.e., undamaged) and straight-frozen neural tissue by modeling each state in 3D. By assessing the degree and type of damage in a straight freeze scenario, we hope to begin building the requisite knowledge base for further, more rapid advances in reconstructive connectomics.
Who knows? Even such “worst case scenarios” may not be as bad as we think.
